Cytoscape was initially released in 2002, and has undergone numerous development sprints. The most recent major sprint ended in 2014 with the release of Cytoscape v3, which refactored major internal data models and greatly improved support for third party plugin applications (via the App Store, semantic versioning disciplines, published APIs, and code management via OSGi).
Since v3, we have focused on addressing outstanding bugs and upgrading a number of user-facing features, including visual styles (VizMapper), fast node filtering, network capacity, and overall speed and UI consistency. The Cytoscape app developer community has upgraded a number of v2 plugins into v3 apps, and has contributed a steady stream of new apps (apps.cytoscape.org).
As of late 2023, our efforts have been rewarded, as Cytoscape has become more popular than ever – 20,000 downloads per month worldwide.
This page explains our roadmap for future deliveries and activities. Feel free to contribute to this discussion our community and user documentation pages.
Cytoscape is going down a number of roads simultaneously:
This page describes the Cytoscape Desktop roadmap and inroads into the wider Cloud Ecosystem.
Cytoscape has traditionally relied on the Cytoscape Apps mechanism for defining and delivering novel calculations and workflows, and the Cytoscape App Store has proven successful at collecting and disseminating them. We are developing Cytoscape Automation features to enable the Cytoscape community to economically create novel workflows using state of the art tools such as Python/Jupyter, R/RStudio and GenomeSpace while leveraging their feature rich libraries and web presence.
Cytoscape Automation will enable users to create workflows executed entirely within Cytoscape or by external tools, and whose results are reproducible. This will enable Cytoscape to scale to large collections of datasets and to larger more complex workflows than is practical via keyboard and mouse.
Cytoscape Automation will exist in two skins – the Commands interface and the Functions interface. Both will accomplish similar results, but will be focused on different usage styles. Commands will reprise user-initiated interactions (e.g., open session, import data, export image), whereas the Functions interface will enable programmers to manipulate and operate on networks as internal Cytoscape data. Commands and Functions will both available via a REST interface.
Under the Cytoscape Automation vision, all new and many existing apps will be upgraded to respond to REST calls. Additionally, most Cytoscape core functions will be exposed via REST calls. In the ideal, all Cytoscape functionality will become controlable from outside workflow systems.
For more information on Cytoscape Automation, consult the Cytoscape Automation wiki.
Before v3.3, we viewed Cytoscape as consisting of the Cytoscape Core as augmented by the Cytoscape Apps. As of v3.3, we have created a middle ground: the Cytoscape Core Apps. The Core is the code that organizes, displays, reads, and writes networks but is agnostic as to network meaning or use – the Core does not contain biology-related functionality. Cytoscape Core Apps are installable modules that perform non-Core functions but are still integral to most Cytoscape usage (e.g., network biology).
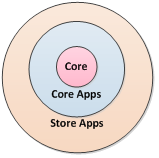
To date, a Cytoscape release has consisted of the Core + Core Apps (Fig. 1). Beginning in v3.3, the Core Apps are available individually in the App Store, though they are all installed with the Core when Cytoscape is installed. However, unlike previous Cytoscape versions, Core Apps are upgradable without releasing all of Cytoscape. (For example, bugs can be fixed or features can be added to the Network Merge at any time.) This enables the Cytoscape team to upgrade specific features much more quickly than in the past.
A close cousin to Core Apps is the App Store concept of Collections, which are bundles of apps that apply to a particular workflow. Installing a Collection causes the installation of all apps in the Collection. From time to time, we will add new Collections or add to existing Collections.
As Cytoscape use cases evolve, we expect to add Core Apps to the Cytoscape release, thereby packaging advanced functionality useful in common workflows. The criteria for promoting an app to a Core App is ultimately decided by the Cytoscape Core developers, but includes:
Additional criteria include:
A top priority for Cytoscape development is to enable efficient execution of new and existing key workflows, including:
Functionality that implements the Core Apps feature is used to enable the user to customize Cytoscape quickly and easily to implement various workflows. Using a Collection (i.e., an app that represents multiple apps), the user can install all apps needed to execute the Functional Enrichment workflow.
| Release | Date | Themes | Features |
3.11 |
TBD 2025 |
|
|
Consequently, we have the following policies, subject to periodic change:
P1: Past releases of Cytoscape Desktop have been made roughly every 12 months, with minor releases for bug fixes and minor features occurring every 3-6 months. Beginning with v3.3, a new version of Cytoscape Desktop is released every six months (~April 15 and ~November 15), with updates of Core Apps to the App Store on an as-needed basis.
P2: A high risk Core feature (which could substantially affect overall usability or which are impossible to definitively verify) must be built into the Core eight weeks before the Core is released, or the feature will be carried over to the next Core release.
P3: Other features must be built into the Core five weeks before Core release.
P4: Bug fixes may be performed (with proper review) up to ten days before Core release.
P5: An external API must be frozen, documented, and with preliminary implementations eight weeks before the Core is released, or the API will be carried over to the next Core release. This includes CyREST functions, parameters and return results. A developer release will be created and made accessible on cytoscape.org.
P6: Milestone builds are created and made available to the user and app developer community eight weeks, five weeks, and ten days before a Core release. If bugs need to be fixed, features need to be withdrawn before the release, or developers see a good reason, additional milestone builds are created.
P7: Milestone builds (including nightly builds) are made available via a developer Download page on the Cytoscape web site.
P8: Emerging API definitions (or references to definitions) are posted on the developer Download page, too.
P9: A preliminary feature list is published five months prior to a Core release, and is updated periodically until ten days before a Core release, at which time it is frozen.
P10: Notably, Cytoscape has expanded beyond its role as a desktop application by also exposing Cytoscape functionality to external control via automation features (also known as Cytoscape Automation). Feature and workflow testing is performed to a large degree through external scripting (e.g., PyUnit tests driven by Python).
P11: Nominations for new Core Apps will be taken until eight weeks before Core release. Core developers can accept prospective Core Apps until five weeks before Core release. Core App nominations not accepted by the this time will be eligible for consideration for the next release.
Note that Core Apps are considered to be part of the Cytoscape core, and are subject to the same Q/A process as core code. Particularly, the interaction of P4 with Core Apps means that changes to Core Apps intended for immediate deployment must undergo review as if they were last minute changes to the core itself. Core Apps will be deployed to the App Store by core developers.
As a member of the Cytoscape Cyberinfrastructure (CI), Cytoscape exchanges networks with a number of CI services, including NDEx and viewers based on cytoscape.js.
One long term vision is to tightly integrate Cytoscape and NDEx so that users naturally load and store their networks using the NDEx service. This enables seamless sharing of networks between Cytoscape and other CI services (e.g., Network Based Stratification), and with other users and publishers. Upon startup, Cytoscape will enumerate the NDEx networks best suited for the user’s use, including networks previously saved by the user and/or the user’s NDEx-based groups, networks pertinent to the user’s interest (e.g., by species, curation properties, etc), or other networks proposed by a recommendation engine. Cytoscape and NDEx will cooperate to make sampling of networks nearly costless so as to encourage exploration (e.g., by caching and displaying network layouts and statistics). Finally, once selected, Cytoscape and NDEx will cooperate to make network loads fast enough that users can remain on task. This vision will initially be realized by a new Core App that replaces the Welcome screen with a panel (a sibling of Network and Select panels) that presents NDEx networks and serves as a jumping off point for workflow execution.
Cytoscape currently exports a network as a file that can be used as supplemental material for a journal article. A journal publisher’s online article viewer is free to display the network in an interactive viewer, which allows the user to zoom into the network and move nodes. In our vision, advanced viewers may perform lookups on selected nodes and show the results in windows, perform new layouts and network analysis/clustering, and export the network to NDEx for integration with Cytoscape and other services – the network would be furnished to the publisher via NDEx directly from Cytoscape instead as a file in supplementary data. Currently, Elsevier offers a simple viewer that accepts a network file and allows zooming and node movement:
The ScienceDirect app makes it very easy for a Cytoscape user to generate supplemental material containing a Cytoscape network, as shown in this movie:
| Version | Release Date | Features | ||||||||||
U24 Award |
||||||||||||
| 3.10.3 | 10/24/2024 |
|
||||||||||
| 3.10.2 | 3/26/2024 |
|
||||||||||
| 3.10.1 | 8/31/2023 |
|
||||||||||
| 3.10.0 | 5/11/2023 |
|
||||||||||
R01 Award (Renewal) |
||||||||||||
| 3.9.1 | 1/24/2022 |
|
||||||||||
| 3.9.0 | 10/21/2021 |
|
||||||||||
| 3.8.2 | 10/29/2020 |
|
||||||||||
| 3.8.1 | 9/24/2020 |
|
||||||||||
| 3.8.0 | 4/17/2020 |
|
||||||||||
| 3.7.2 | 9/5/2019 |
|
||||||||||
| 3.7.1 | 1/10/2019 |
|
||||||||||
| 3.7.0 | 10/22/2018 |
|
||||||||||
| 3.6.1 | 3/26/2018 |
|
||||||||||
| 3.6 | 11/15/2017 |
|
||||||||||
| 3.5.1 | 4/26/2017 |
|
||||||||||
| 3.5 | 3/24/2017 |
|
||||||||||
| 3.4 | 05/13/2016 |
|
||||||||||
| 3.3 | 11/25/2015 |
|
||||||||||
| 3.2.1 | 02/11/2015 |
|
||||||||||
| 3.2 | 11/07/2014 |
|
||||||||||
| 3.1.1 | 05/27/2014 |
|
||||||||||
| 3.1 | 02/13/2014 |
|
||||||||||
| 3.0.2 | 07/30/2013 |
|
||||||||||
R01 Award (Initial) |
||||||||||||
| 3.0.1 | 04/22/2013 |
|
||||||||||
| 3.0 | 02/04/2013 |
|
||||||||||
| 2.8.3 | 05/04/2012 |
|
||||||||||
| 2.8.2 | 08/26/2011 |
|
||||||||||
| 2.8.1 | 02/18/2011 |
|
||||||||||
| 2.8.0 | 12/2/2010 |
|
||||||||||
| 2.7.0 | 04/27/2010 |
|
||||||||||
| 2.6.3 | 06/25/2009 | |||||||||||
| 2.6.2 | 02/24/2009 | |||||||||||
| 2.6.1 | 09/17/2008 | |||||||||||
| 2.6.0 | 05/15/2008 | |||||||||||
| 2.5.2 | 02/01/2008 | |||||||||||
| 2.5.1 | 08/31/2007 | |||||||||||
| 2.5.0 | 07/23/2007 | |||||||||||
| 2.4.1 | 04/25/2007 | |||||||||||
| 2.4.0 | 01/16/2007 | |||||||||||
| 2.3.2 | 09/01/2006 | |||||||||||
| 2.3.1 | 07/19/2006 | |||||||||||
| 2.3 | 06/23/2006 | |||||||||||
| 2.2 | 12/06/2005 | |||||||||||
| 2.1 | 05/25/2005 | |||||||||||
| 2.0 | 07/22/2004 | |||||||||||
| 1.1 | May 2003 | |||||||||||
| 1.0 | Mar 2003 | |||||||||||
| 0.9 | Nov 2002 | |||||||||||
| 0.8 | Jul 2002 | Release notes for all prior and subsequent releases can be found here |